Large-scale evolutionary analysis of the transcriptome of poison arrow frogs
Introduction
The transcriptome (the sum total of all the messenger RNA molecules expressed from the genes of an organism) of poison arrow frogs has not been studied before. There are nocturnal and diurnal species of poison arrow frog, that differ greatly in appearance (Figure 1) making them an ideal subject for studying niche adaptation.
Evidence from mammal and snake studies indicate that genetic differences in visual genes are associated with adaptation to light. To date, amphibia have not been studied. In this project we studied nocturnal and diurnal poison arrow frogs to identify gene expression differences and to identify protein-coding sequences for future analysis for molecular adaptation studies.

Figure 1. Examples of diurnal (A) and nocturnal (B) poison arrow frogs
Data and methods
The data for this project consisted of RNA-Seq data obtained from RNA extracted from the eyes of 8 different poison arrow frogs species (Table 1).
Table 1. Sample information
| Species | Activity | Number of Samples |
| Conspicuus | Diurnal | 4 |
| Lamasi | Diurnal | 2 |
| Perezi | Diurnal | 1 |
| Trivittata | Diurnal | 2 |
| Parviceps | Nocturnal | 2 |
| Royi | Nocturnal | 4 |
| Ruber | Nocturnal | 2 |
| Sp | Nocturnal | 3 |
Data processing was completed using the marc1 HPC facility at the University of Leeds. An overview of the analysis pipeline devised for data processing, quality control and analysis is shown in figure 2.
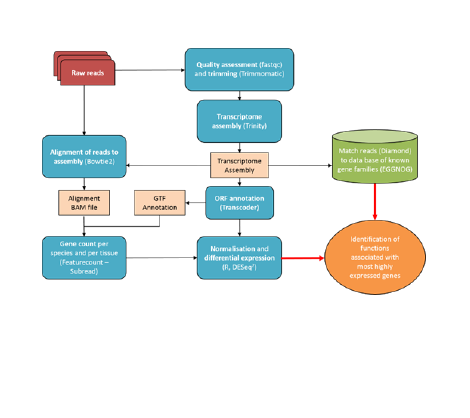
Figure 2. Planned Analysis Pipeline
Key findings
Quality assessment of the RNA-Seq data using fastqc has established that the sequence quality is very high, with all raw files demonstrating a per base sequence quality of at least 99.99%. Adapter contamination was evident, however, these sequences were trimmed prior to transcriptome assembly.
Transcriptomes were successfully assembled for all 8 PAF species studied.
Value of the research
Research into non-model species is lagging far behind – projects such as this provide a unique opportunity to generate data for the community for important non-model systems. For example, the assembled transcriptomes can be used as a proxy for the protein-coding genome. As a result, it will be possible to investigate differential gene expression in poison arrow frogs eyes. Amphibia represent a major transition in animal evolution from aquatic to terrestrial life, with studies from terrestrial species such as mammals and snakes indicating changes in expression of key visual genes, it is imperative that amphibia are also studied to better understand the evolution of vision in the animal kingdom.
Researchers
Katie Nicoll Baines - Leeds Institute for Data Analytics, and Computational and Molecular Evolutionary Biology Group, University of Leeds,
Karen Siu Ting - Computational and Molecular Evolutionary Biology Group, University of Leeds & Institute of Biological, Environmental and Rural Sciences, Aberystwyth University
Chris J. Creevey - Institute of Biological, Environmental and Rural Sciences, Aberystwyth University
Mary J. O’Connell - Computational and Molecular Evolutionary Biology Group, University of Leeds
